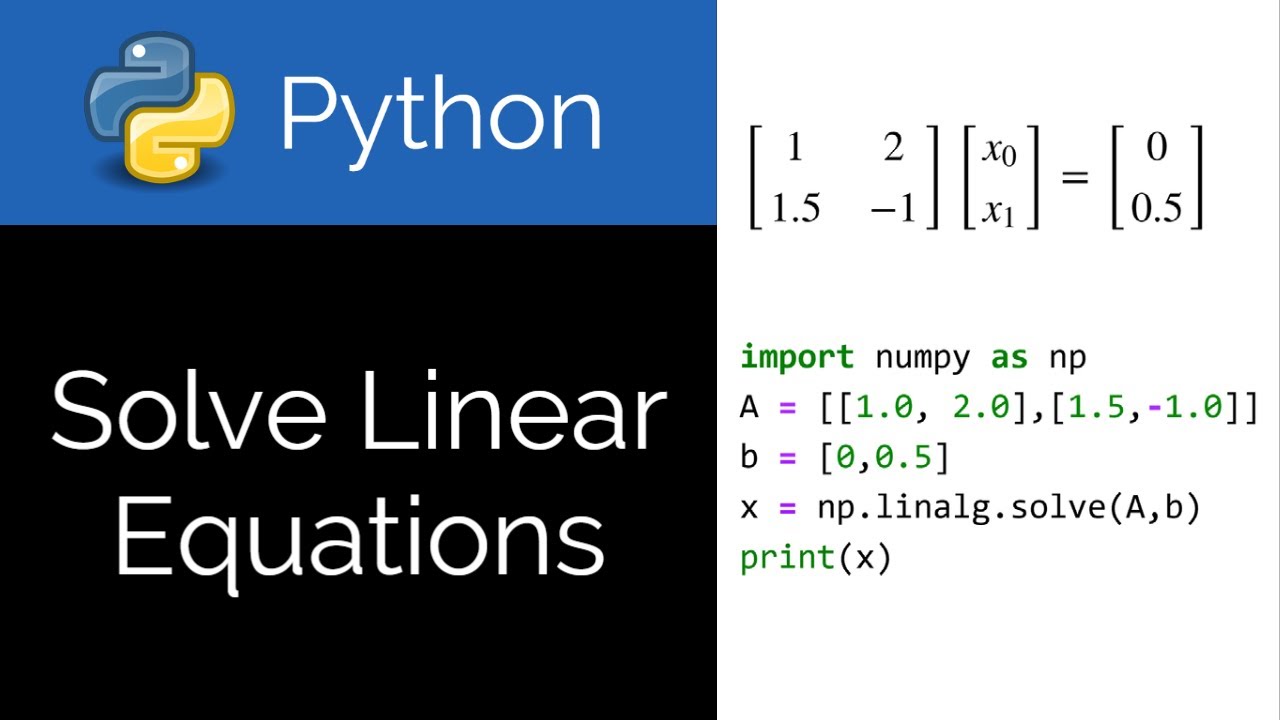
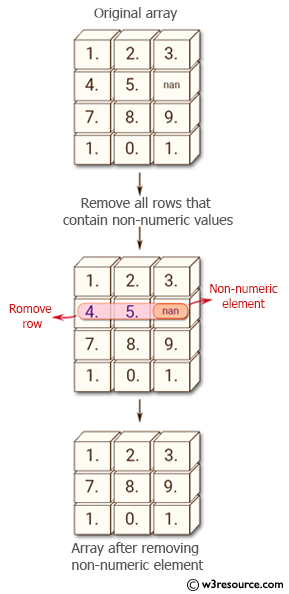
Numpy loadtxt numpy loadtxt fname dtype class float comments delimiter none converters none skiprows 0 usecols none unpack false ndmin 0 encoding bytes max rows none source load data from a text file.
Read mat file in python numpy.
I have a data in mat file observations and features and i want to load it into numpy 2d array.
From scipy io import loadmat annots loadmat cars train annos mat well it s really that simple.
This can be disabled by setting the optional argument struct as record false.
Beginning at release 7 3 of matlab mat files are actually saved using the hdf5 format by default except if you use the vx flag at save time see in matlab.
Parameters fname file str or pathlib path.
Matlab writes matrices in fortran order so this also transposes matrices and higher dimensional arrays into conventional numpy order arr page row col.
Reading them in is definitely the easy part.
File filename or generator to read.
These files can be read in python using for instance the pytables or h5py package.
You can get it done in one line of code.
The read method returns the specified number of bytes from the file.
Scipy is a really popular python library used for scientific computing and quite naturally they have a method which lets you read in mat files.
Each row in the text file must have the same number of values.
Default is 1 which means the whole file.
To load a csv comma separated values file we specify delimitter to.
A cheap way to save out data for matlab within python is as follows.
I dont want to convert it into csv first and then load csv into numpy.
Numpy scipy scikit learn opencv python.
Numppy s loadtxt function lets us read numerical data file in text format in to python.
By default scipy reads matlab structs as structured numpy arrays where the dtype fields are of type object and the names correspond to the matlab struct field names.
Reading matlab structures in mat files does not seem supported at this point.
Suppose you have a numpy array of shape 10777 9 called pkt np array.